Has anyone used Pylinac for end to end testing? Is there a module that I could start with that would be able to identify gafchromic pinprick points? Maybe there are other tools in python that could work with Pylinac also.
Landon
Has anyone used Pylinac for end to end testing? Is there a module that I could start with that would be able to identify gafchromic pinprick points? Maybe there are other tools in python that could work with Pylinac also.
Landon
Does your need allow you to give an estimate first? Or does it need to bootstrap everything? Generally, I’ve found that if you can grab/crop down to the approximate region and then perform a sum of each axis you can find your peak/valley relatively robustly. Then again, I’ve purposely not supported film for a reason. You probably already know about the image module, but for outside readers there is an image module in pylinac for doing more basic operations (load, crop, roll, filter, …): Core Modules Documentation — pylinac 2.4.0 documentation
My use case would be running from QATrack ideally. I haven’t analyzed any film images so I’m not really sure where to start though. I didn’t know how difficult it would be, its just one of the tests I have yet to add. Thanks
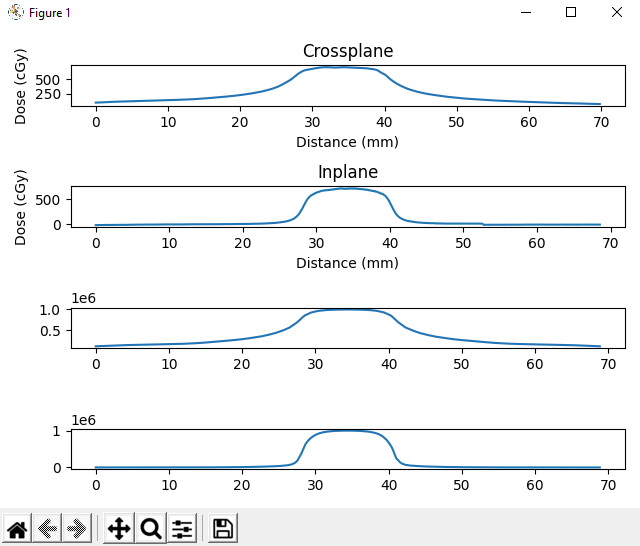
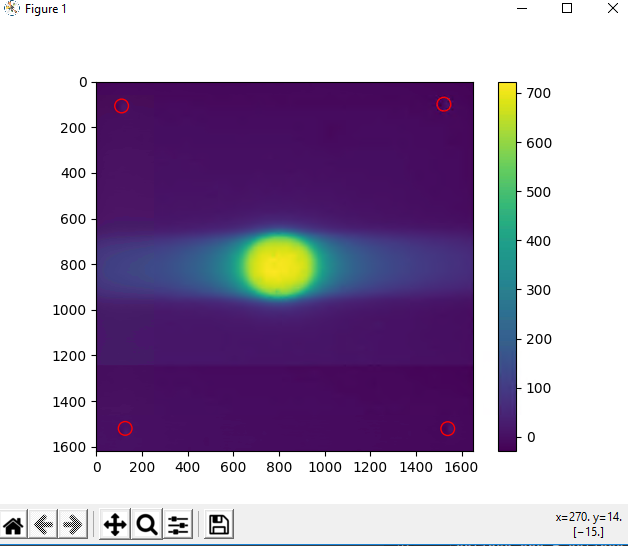
This is sort of what I was thinking - I had to use tifffile to import the green portion of the gafchromic tiff file. I used gaussian filtering for smoothing (is there a better method for smoothing noisy film?) Then I used the pylinac.ct module to find the film pinholes (they are spaced 6 cm apart) and retrieved profiles from the center of the 4 pinholes. The dose from the film (top two plots) look about what I was thinking absolute dose wise. The bottom two were plotted from the dicom dose file loaded from pydicom. The majority of the math performed on the film was from “Practical IMRT QA dosimetry using Gafchromic film: a quick start guide”.
I can’t figure out how to get absolute dose from the Eclipse RD.dcm file…does pylinac have a method to find absolute dose scaling from a dicom file? It seems to be normalizing to 1. Probably not the most robust python file to get results, but it would be nice to periodically do end to end tests without spending a lot of time on analysis.
Nice work Landon.
Re: filters. Pylinac’s filter options are gaussian and median. I usually prefer median since they are edge-preserving. It’s just a wrapper of scipy so feel free to dig into scipy’s filtering options. Better yet, scikit-image does even more.
Re: dose. Make sure you’ve exported the dose in absolute format vs relative. Then to get the absolute dose multiply the dose ndarray by the DoseGridScaling tag. For me, this gives the absolute dose in Gy.
Generally speaking, I hate film so it’s unlikely a tool like this will come about in fully-fleshed out form. However, I am working on making pylinac more flexible (again, generally speaking) which would include some tools like finding a pin prick, edge, etc given an ROI.