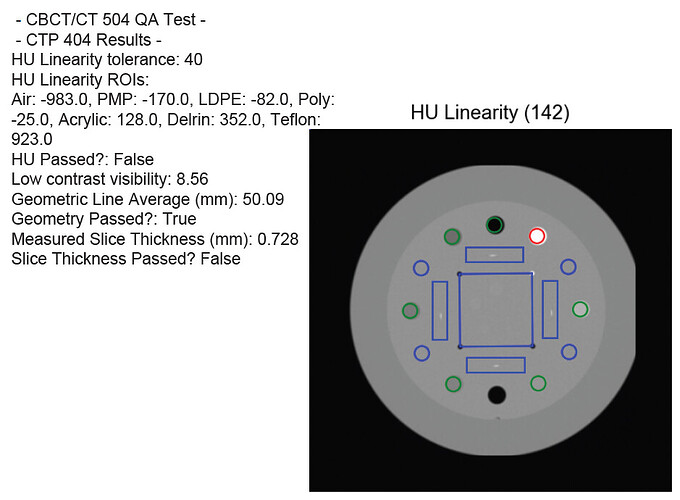
Dear pylinac team,
I’m new to pylinac and I’m trying to analyze dicom images of a catphan 504. Here’s the snippet:
from pathlib import Path
from pylinac import CatPhan504
cbct_folder = Path(“path/to/folder”)
mycbct = CatPhan504(cbct_folder)
mycbct.analyze()
But I get the following error message:
TypeError Traceback (most recent call last)
Cell In[40], line 1
----> 1 mycbct.analyze()
File ~\fss\fss\Lib\site-packages\pylinac\ct.py:2806, in CatPhanBase.analyze(self, hu_tolerance, scaling_tolerance, thickness_tolerance, low_contrast_tolerance, cnr_threshold, zip_after, contrast_method, visibility_threshold, thickness_slice_straddle, expected_hu_values, x_adjustment, y_adjustment, angle_adjustment, roi_size_factor, scaling_factor, origin_slice, roll_slice_offset)
2804 self.scaling_factor = scaling_factor
2805 self.roll_slice_offset = roll_slice_offset
→ 2806 self.localize(origin_slice)
2807 ctp404, offset = self._get_module(CTP404CP504, raise_empty=True)
2808 self.ctp404 = ctp404(
2809 self,
2810 offset=offset,
(…) 2816 expected_hu_values=expected_hu_values,
2817 )
File ~\fss\fss\Lib\site-packages\pylinac\ct.py:2356, in CatPhanBase.localize(self, origin_slice)
2354 def localize(self, origin_slice: int | None) → None:
2355 “”“Find the slice number of the catphan’s HU linearity module and roll angle”“”
→ 2356 self._phantom_center_func = self.find_phantom_axis()
2357 if origin_slice is not None:
2358 self.origin_slice = origin_slice
File ~\fss\fss\Lib\site-packages\pylinac\ct.py:2440, in CatPhanBase.find_phantom_axis(self)
2435 common_idxs = np.intersect1d(x_idxs, y_idxs)
2436 # fit to 1D polynomials; inspiration: https://stackoverflow.com/a/45351484
2437 # rcond should be explicitly passed. Started randomly failing in the pipe. v1.14.0 numpy release notes
2438 # say it should be explicitly passed. Value is arbitrary but small and tests pass.
2439 fit_zx = np.poly1d(
→ 2440 np.polyfit(zs[common_idxs], center_xs[common_idxs], deg=1, rcond=0.00001)
2441 )
2442 fit_zy = np.poly1d(
2443 np.polyfit(zs[common_idxs], center_ys[common_idxs], deg=1, rcond=0.00001)
2444 )
2445 return fit_zx, fit_zy
File ~\fss\fss\Lib\site-packages\numpy\lib_polynomial_impl.py:645, in polyfit(x, y, deg, rcond, full, w, cov)
643 raise TypeError(“expected 1D vector for x”)
644 if x.size == 0:
→ 645 raise TypeError(“expected non-empty vector for x”)
646 if y.ndim < 1 or y.ndim > 2:
647 raise TypeError(“expected 1D or 2D array for y”)
TypeError: expected non-empty vector for x
I know, as a matter of fact that the image is a bit clipped as it can be seen in the figure attached.
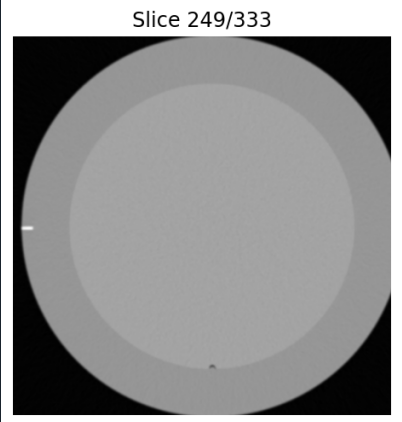
I was wondering if that’s really what is causing the error message, since the clipping doesn’t seem to be too much. Also, if that’s the case, if there’s any workaround for this issue.
I can also provide the dicom file if that’s useful. If the problem is not the clipping, which dicom parameters should I look at and correct?
Huge thanks and thanks in advance for your reply.
Best wishes,
Gustavo