I’m having trouble figuring out how to adjust the CatPhan 503 modules. It looks like I’m about a slice off or two. I’m assuming I would have to adjust the class pylinac.ct.slice with the slice_num parameter to get the correct slice.
Hi,
You could adjust the module locations slightly by making a custom class (copied from CatPhan/CT — pylinac 3.0.2 documentation):
Similar to partial scans, to modify the module location(s), overload the modules attr and edit the offset value. The value is in mm:
from pylinac import CatPhan504 # works for any of the other phantoms too
from pylinac.ct import CTP515, CTP486, CTP528
create custom catphan with module locations
class OffsetCatPhan504(CatPhan504):
modules = {
CTP486: {‘offset’: -60}, # normally -65
CTP528: {‘offset’: 30},
CTP515: {‘offset’: -25}, # normally -30 }
ct = OffsetCatPhan504.from_zip(…) # use like normal
Cheers,
Tim
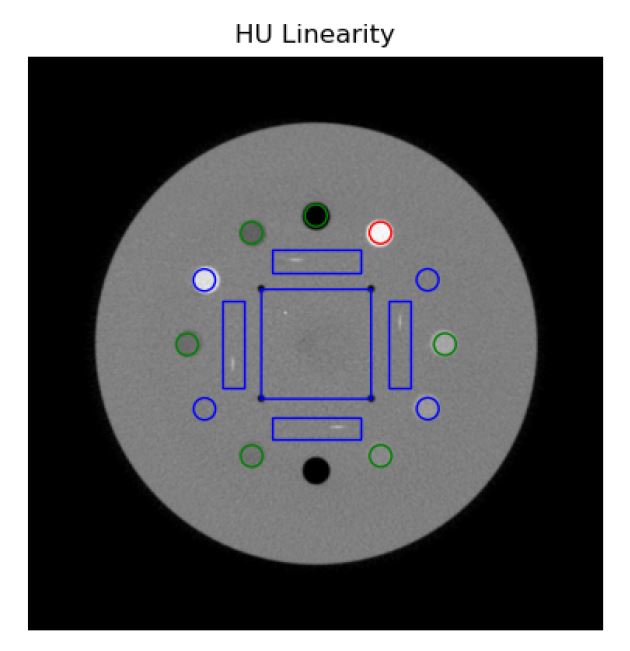

Tim is correct. The documentation covers this use case: CatPhan/CT — pylinac 3.0.2 documentation. Here’s a gist: https://gist.github.com/jrkerns/f1ee9e80ea04af375c57640b84315daa


