Hi,
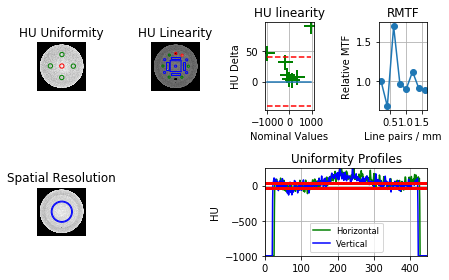
I’m analyzing a Catphan 503, from a Gamma Knife CBCT. The slice spacing is 0.5mm. Pylinac doesn’t seem to find the spatial resolution section properly? See below:
James, can I send some sample data to see if you can reproduce?
Many thanks,
Ben
Hi,
I’m analyzing a Catphan 503, from a Gamma Knife CBCT. The slice spacing is 0.5mm. Pylinac doesn’t seem to find the spatial resolution section properly? See below:

James, can I send some sample data to see if you can reproduce?
Many thanks,
Ben
Hey Ben,
Yes please!
Hi Ben,
When I ran the files it didn’t even find the HU module. After some digging I saw the problem: the images are basically much noisier than pylinac is expecting and threw out all or some of the HU module slices. The culprit is here: https://github.com/jrkerns/pylinac/blob/master/pylinac/ct.py#L1123. This expression is saying: “The HU module should have an extreme high (teflon), an extreme low (air), and the majority of it should be uniform (percentiles).” Unfortunately, due to the noise the percentile measure is failing. When set to 200 some of the HU module was picked up and I got your results. When set to 250 everything worked as normal. For the short term, I would simply change that value in your local install. I need to run my bank of tests to see if changing this may have an effect elsewhere for other datasets.